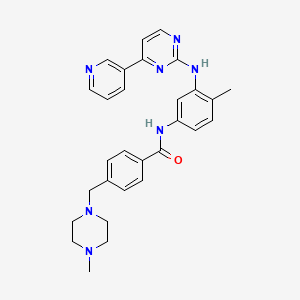
D1377 | Imatinib
L
L01XE01 Imatinib
[L01XE] Protein kinase inhibitors
[L01X] OTHER ANTINEOPLASTIC AGENTS
[L01] ANTINEOPLASTIC AGENTS
[L] Antineoplastic and immunomodulating agents
| Toxicity | Dose | Time | Species | Model | Method | Action | Positive criterion | Reference |
|---|---|---|---|---|---|---|---|---|
| MEMBRANE POTENTIAL | 25.7 µM | 30 mins | mouse | liver mitochondria | Rh123 fluorescence (excitation 485 nm, emission 535 nm) are recorded using a fluorescence multi-well plate reader (mCICCP (20 µM) treatments was considered as the 100% baseline for ΔΨm loss) | decrease | EC20 | 36 |
| RESPIRATION | ND | 60 mins | mouse | liver mitochondria | Oxygen consumption was monitored with 50nM MitoXpress ( an oxygen-sensitive phosphorescent dye) using a spectrofluorimeter (Tecan Infinite 200; λExcitation 380nm; λEmission 650nm). Rotenone (2µM) was used as 100% baseline for complex I inhibition. | Negative | EC20 | 36 |
| RESPIRATION | ND | 60 mins | mouse | liver mitochondria | Oxygen consumption was monitored with 50nM MitoXpress ( an oxygen-sensitive phosphorescent dye) using a spectrofluorimeter (Tecan Infinite 200; λExcitation 380nm; λEmission 650nm). Oligomycin A (1µM) was used as 100% baseline for complex II inhibition. | Negative | EC20 | 36 |
| GLYCOLYSIS | 249 | |||||||
| SWELLING | ND | 30 mins | mouse | liver mitochondria | swelling assay: Absorbance at 545 nm using a fluorescence multi-well plate reader (CaCl2 (50 µM) was considered as the 100% baseline for the swelling ) | Negative | EC20 | 36 |
| Target | Dose | Time | Species | Model | Method | Action | Positive criterion | Reference |
|---|---|---|---|---|---|---|---|---|
| NADH:ubiquinone reductase | ND | 60 mins | mouse | liver mitochondria | Oxygen consumption was monitored with 50nM MitoXpress ( an oxygen-sensitive phosphorescent dye) using a spectrofluorimeter (Tecan Infinite 200; λExcitation 380nm; λEmission 650nm). Rotenone (2µM) was used as 100% baseline for complex I inhibition. | Negative | EC20 | 36 |
| Succinate dehydrogenase | ND | 60 mins | mouse | liver mitochondria | Oxygen consumption was monitored with 50nM MitoXpress ( an oxygen-sensitive phosphorescent dye) using a spectrofluorimeter (Tecan Infinite 200; λExcitation 380nm; λEmission 650nm). Oligomycin A (1µM) was used as 100% baseline for complex II inhibition. | Negative | EC20 | 36 |
| Cytochrome c | 163.6 µM | 30 mins | mouse | liver mitochondria | Cytochrome c release was evaluated using ELISA kit ( 20 µg/ml Alamethicin was used as 100% baseline) | release | EC20 | 36 |
| Pictogram | Signal | Statements | Precautionary Statement Codes |
|---|---|---|---|
   |
Danger |
Aggregated GHS information provided by 9 companies from 6 notifications to the ECHA C&L Inventory. Each notification may be associated with multiple companies. H302 (22.22%): Harmful if swallowed [Warning Acute toxicity, oral] H315 (11.11%): Causes skin irritation [Warning Skin corrosion/irritation] H319 (11.11%): Causes serious eye irritation [Warning Serious eye damage/eye irritation] H341 (55.56%): Suspected of causing genetic defects [Warning Germ cell mutagenicity] H351 (77.78%): Suspected of causing cancer [Warning Carcinogenicity] H360 (55.56%): May damage fertility or the unborn child [Danger Reproductive toxicity] H361 (33.33%): Suspected of damaging fertility or the unborn child [Warning Reproductive toxicity] H362 (55.56%): May cause harm to breast-fed children [Reproductive toxicity, effects on or via lactation] H373 (22.22%): Causes damage to organs through prolonged or repeated exposure [Warning Specific target organ toxicity, repeated exposure] H411 (22.22%): Toxic to aquatic life with long lasting effects [Hazardous to the aquatic environment, long-term hazard] Information may vary between notifications depending on impurities, additives, and other factors. The percentage value in parenthesis indicates the notified classification ratio from companies that provide hazard codes. Only hazard codes with percentage values above 10% are shown. |
P201, P202, P260, P263, P264, P270, P273, P280, P281, P301+P312, P302+P352, P305+P351+P338, P308+P313, P314, P321, P330, P332+P313, P337+P313, P362, P391, P405, and P501; (The corresponding statement to each P-code can be found at the GHS Classification page.) |
| 1080014-82-9 | 112GI019 | 12810-EP2269994A1 |
| 12810-EP2270008A1 | 12810-EP2272827A1 | 12810-EP2275412A1 |
| 12810-EP2275413A1 | 12810-EP2277865A1 | 12810-EP2287156A1 |
| 12810-EP2289892A1 | 12810-EP2291366A2 | 12810-EP2292234A1 |
| 12810-EP2292617A1 | 12810-EP2305667A2 | 12810-EP2308855A1 |
| 12810-EP2311807A1 | 12810-EP2311821A1 | 12810-EP2311840A1 |
| 12810-EP2316831A1 | 12810-EP2316832A1 | 12810-EP2316833A1 |
| 152459-95-5 | 1iep | 1xbb |
| 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE | 4-[(4-Methyl-1-piperazinyl)methyl]-N-[4-methyl-3-[[4(3-pyridinyl)-2-pyrimidinyl]amino]phenyl]-benzamide | 4-[(4-Methyl-1-piperazinyl)methyl]-N-[4-methyl-3-[[4-(3- pyridinyl)-2-pyrimidinyl]amino]phenyl]benzamide |
| 4-[(4-Methyl-1-piperazinyl)methyl]-N-[4-methyl-3-[[4-(3-pyridinyl)-2-pyrimidinyl]amino]-phenyl]benzamide | 4-[(4-methyl-1-piperazinyl)-methyl]-N-{4-methyl-3-[[4-(3-pyridinyl)-2-pyrimidinyl]-amino]-phenyl)-benzamide | 4-[(4-methyl-1-piperazinyl)-methyl]-N-{4-methyl-3-[[4-(3-pyridinyl)-2-pyrimidinyl]-amino]-phenyl}-benzamide |
| 4-[(4-methyl-1-piperazinyl)methyl]-N-[4-methyl-3-[(4-(3-pyridinyl)-2-pyrimidinyl]amino]-phenyl]benzamide | 4-[(4-methyl-1-piperazinyl)methyl]-N-[4-methyl-3-[[4-(3-pyridinyl)-2-pyrimidinyl]amino]phenyl]-benzamide | 4-[(4-methyl-1-piperazinyl)methyl]-N-[4-methyl-3-[[4-(3-pyridinyl)-2-pyrimidinyl]amino]phenyl]-benzamide methanesulfonate |
| 4-[(4-methyl-1-piperazinyl)methyl]-N-[4-methyl-3-[[4-(3-pyridinyl)-2-pyrimidinyl]amino]phenyl]benzamide | 4-[(4-methyl-1-piperazinyl)methyl]-n[4-methyl-3-[[4-(3-pyridinyl)-2-pyrimidinyl]amino]phenyl]-benzamide | 4-[(4-methylpiperazin-1-yl)methyl]-N-(4-methyl-3-{[4-(pyridin-3-yl)pyrimidin-2-yl]amino}phenyl)benzamide |
| 4-[(4-methylpiperazin-1-yl)methyl]-N-[4-methyl-3-[(4-pyridin-3-ylpyrimidin-2-yl)amino]phenyl]benzamide | 4-[(4-methylpiperazin-1-yl)methyl]-N-{4-methyl-3-[(4-pyridin-3-ylpyrimidin-2-yl)amino]phenyl}benzamide | 459I955 |
| AB0044023 | AB00698388-07 | AB00698388-10 |
| AB00698388-11 | AB00698388-12 | AB00698388-13 |
| AB00698388_15 | AB00698388_16 | AC-524 |
| ACMC-20a8ej | AK107630 | AKOS000280662 |
| AM20090646 | ANW-61817 | API0024674 |
| AX8134347 | BCP01542 | BCP9000775 |
| BCPP000205 | BDBM13530 | BIDD:GT0047 |
| BKJ8M8G5HI | BRD-K92723993-066-02-9 | BRD-K92723993-066-04-5 |
| Benzamide, 4-((4-methyl)-1-piperazinyl)methyl)-N-(4-methyl-3-((4-(3-pyridinyl)-2-pyrimidinyl)amino)phenyl)- | Benzamide, 4-[(4-methyl-1-piperazinyl)methyl]-N-[4-methyl-3-[[4-(3-pyridinyl)-2-pyrimidinyl]amino]phenyl]- (9CI) | Benzamide, 4-[(4-methyl-1-piperazinyl)methyl]-N-[4-methyl-3-[[4-(3-pyridinyl)-2-pyrimidinyl]amino]phenyl]-, methanesulfonate (1:1);4-[(4-Methyl-1-piperazinyl)methyl]-N-[4-methyl-3-[[4-(3-pyridinyl)-2-pyrimidinyl]amino]phenyl]-benzamide monomethanesulfonate;imatinib;Imatinib mesylate |
| C29H31N7O | CCG-101289 | CCRIS 9076 |
| CGP 57148B | CGP-57148 | CHEBI:45783 |
| CHEMBL941 | CS-0964 | CTK8B9010 |
| Cgp 57148 | D08066 | DB00619 |
| DTXSID3037125 | ES-0058 | EX-A063 |
| FT-0651483 | GTPL5687 | Glamox (TN) |
| Gleevec (TN) (Novartis) | Gleevec(TM) | Glivec |
| HMS2089D03 | HMS3244P06 | HMS3244P10 |
| HMS3244P14 | HMS3656K04 | HMS3715P03 |
| HY-15463 | I0906 | IMATINIB |
| Imatinib | Imatinib (Gleevec) | Imatinib (INN) |
| Imatinib (STI571) | Imatinib - Gleevec | Imatinib Methansulfonate |
| Imatinib [INN:BAN] | Imatinib free base | Imatinib, 21 |
| KS-00000GF2 | KSC919A1B | Kinome_3724 |
| LS-182208 | LS-187106 | MCULE-2384256888 |
| MRF-0000449 | N-(3-(4-(pyridin-3-yl)pyrimidin-2-ylamino)-4-methylphenyl)-4-((4-methylpiperazin-1-yl)methyl)benzamide | N-(4-Methyl-3-((4-(pyridin-3-yl)pyrimidin-2-yl)amino)phenyl)-4-((4-methylpiperazin-1-yl)methyl)benzamide |
| N-(4-methyl-3-(4-(pyridin-3-yl)pyrimidin-2-ylamino)phenyl)-4-((4-methylpiperazin-1-yl)methyl)benzamide | N-(4-methyl-3-{[4-(pyridin-3-yl)pyrimidin-2-yl]amino}phenyl)-4-[(4-methylpiperazin-1-yl)methyl]benzamide | NCGC00159456-02 |
| NCGC00159456-03 | NCGC00159456-04 | NCGC00159456-05 |
| NCGC00159456-06 | NCGC00159456-07 | NCGC00159456-09 |
| NCGC00159456-16 | NS00009172 | NSC-743414 |
| NSC-759854 | NSC743414 | NSC759854 |
| Pharmakon1600-01502276 | Q-201231 | Q177094 |
| QCR-269 | S2475 | SB17306 |
| SCHEMBL3827 | SR-01000763561 | SR-01000763561-4 |
| SR-01000763561-6 | ST 1571 | ST1571 |
| STI | STI 571 | STI-571 |
| STI571 | STK617705 | SW197805-5 |
| UNII-BKJ8M8G5HI | VCC905240 | W-3956 |
| Z1551429727 | ZINC19632618 | alpha-(4-Methyl-1-piperazinyl)-3'-((4-(3-pyridyl)-2-pyrimidinyl)amino)-p-tolu-p-toluidide |
| alpha-(4-methyl-1-piperazinyl)-3'-((4-(3-pyridyl)-2-pyrimidinyl)amino)-p-toluidide | benzamide, 4-[(4-methyl-1-piperazinyl)methyl]-N-[4-methyl-3-[[4-(3-pyridinyl)-2-pyrimidinyl]amino]phenyl]- | cid_5291 |
| imatinib-CD3 | sti-571 |